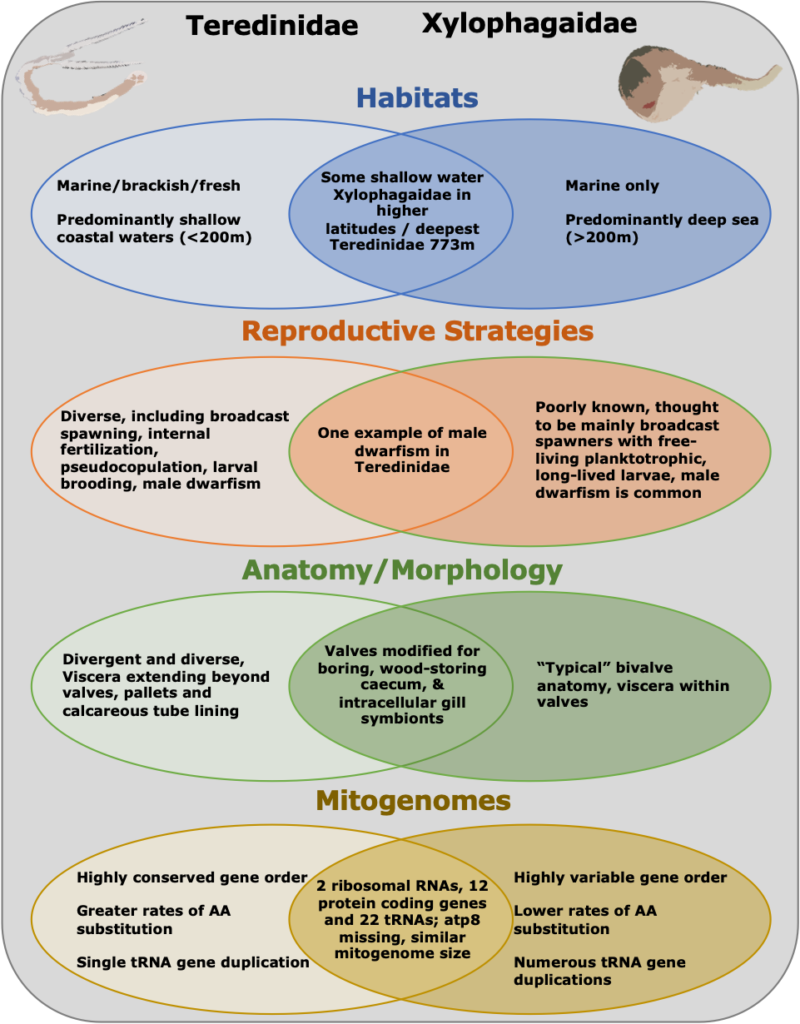
You might know that many animals in the deep sea evolved from shallow water ancestors, but did you ever wonder what happens to their genes when they make that evolutionary trip? Researchers at OGL asked that question by looking at the mitochondria of two families of marine clams: one that stayed in shallow water, and a closely related sister family that, over many millions of years, migrated into the deep.
Mitochondria are tiny bean-shaped structures found in cells of all animals. They are known as the powerhouse of the cell because they help convert food into the fuel (ATP) that cells need to survive. Surprisingly, the mitochondria have their own tiny genomes, separate from that of the rest of the cell. The small size of mitochondrial genomes makes them a quick read, and that makes them attractive for scientists to study.
The question the researchers asked was: How did their mitochondrial genes change when these clams moved from the warm, food-rich, shallow ocean, to the cold, food-poor desert of the deep sea? The answer was unexpected. The genes in the deep-sea family became shuffled, with different gene orders in different species, but the rate at which the individual genes evolved was slow. In the shallow water family, the genes evolved quickly, but they all stayed in the same order. In the future, the researchers hope to discover why these two families followed such different evolutionary paths and what this might tell us about life in the deep sea.
Read more about this fascinating work here.
Interested in helping OGL dive into more marine genomes? Support OGL here.
