Ever wonder how scientists preserve DNA samples for groundbreaking research? When researchers collect tissue samples in the field, it’s really important to preserve the DNA in the tissue, otherwise it quickly breaks down. Once DNA begins to degrade, it becomes more difficult to analyze, so researchers rely on freezing or various liquid preservatives to protect samples once they are collected.
Surprisingly, while DNA sequencing methods have improved by leaps and bounds, preservation methods have not changed much in 100 years. In fact, the most common preservatives in use today are the same ones used by Charles Darwin and his contemporaries!
Nonetheless, some newer preservative solutions are gaining traction. To test them, OGL has launched a new long-term undergraduate student research program called “Student Research in DNA Preservation.” The program employs undergraduate student interns in a continuous daisy chain of connected six-month projects to tackle the long-term research needed to evaluate biological preservatives.

We’re especially proud of this program because three of our undergraduate co-op interns—Amy Sharpe, Sonia Barrios, and Sarah Gayer—are now co-authors on a recently published paper from OGL! Undergraduate research is critical to our work at OGL, and each of our co-ops have made important contributions to studying DNA preservation. This not only helps with our research, but also gives Northeastern undergraduates the opportunity to learn about each step of the research process along with the potential to author a paper!
We are excited to announce the first paper resulting from this project, which seeks to better understand how each of the three ingredients in an increasingly popular preservative called DMSO-salt contribute to DNA preservation. The ingredients of this preservative are a solvent called DMSO, table salt (NaCl), and a common food additive called EDTA. While it is reasonable to predict that all three of these ingredients could contribute to DNA preservation, our study shows that only EDTA does! This means that when researchers preserve tissue samples in EDTA, a significant amount of DNA remains intact so that researchers can analyze it. This is an important discovery because not only is EDTA very effective at preservation, but because it is a common food preservative, there is little risk to researchers when handling it.
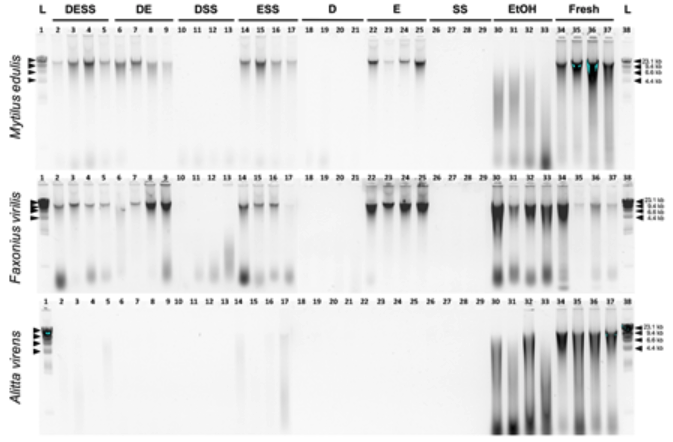
OGL continues to study ways to improve the preservation of DNA. This research is important to ensure that scientists have better options for preserving samples while in the field. Instead of having to lug around heavy containers of liquid nitrogen to flash freeze samples, they can carry small quantities of a non-hazardous solution! Our hope is that our research will improve the quality of genetic samples collected around the world, so that the important work of preserving the ocean’s genome will become easier for researchers everywhere.
You can read the full published paper here to learn more about OGL’s research!
Interested in helping to support the Student Research in DNA Preservation project? Support OGL’s research here!
